Medical imaging is an important part of modern medicine, and the ability to process and analyze medical images is crucial for diagnosis, treatment, and research. One of the most common formats for storing medical images is DICOM (Digital Imaging and Communications in Medicine), a standard used by medical professionals to store and exchange medical images and related information. PyDICOM is a Python package for working with DICOM files, and in this article, we will explore its features and how to use it to work with medical images in Python.
Installing PyDICOM Package
To start working with PyDICOM, you first need to install it in python. You can install it using pip (Python package manager) by running the following command. PyDICOM also requires additional packages like Pillow, libjpeg for pixel data processing. Those will be automatically installed with PyDICOM installation.
pip install pydicomOnce PyDICOM is installed, you can begin working with DICOM files processing in Python. You can load any DICOM file and read its content and metadata.
Reading DICOM Files
The first step in working with DICOM files is to read them using PyDICOM. PyDICOM provides a simple API for reading DICOM files. You can read a DICOM file by creating a Dataset object and passing the path to the file as an argument. Here is an example:
import pydicom
# read from DICOM file.
ds = pydicom.dcmread('image.dcm')In this example, we read a DICOM file named image.dcm and store it in the ds variable. The dcmread() function returns a Dataset object that represents the DICOM file. This Dataset object contains all the information stored in the DICOM file, including image data, metadata, and patient information
Accessing Metadata
Once you have read a DICOM file, you can access its metadata using the Dataset object. The metadata contains information such as patient name, study date, image size, and more. You can access metadata by using the names of the DICOM tags. Here is an example:
patient_name = ds.PatientName
study_date = ds.StudyDate
image_size = (ds.Rows, ds.Columns)
print("Patient Name:", patient_name)
print("Studay Date:", study_date)
print("Image Size:", image_size)Patient Name: Anonymized
Studay Date: 20010109
Image Size: (2022, 2006)In this example, we access the patient name, study date, and image size metadata and store them in variables. Note that the names of the DICOM tags are case sensitive.
Accessing Image Data
In addition to metadata, DICOM files also contain image data. You can access image data using the pixel_array attribute of the Dataset object. In some case, you may need to install additional packages like libjpeg for pixel data processing. Here is an example:
pip install pylibjpeg-libjpegimport matplotlib.pyplot as plt
image = ds.pixel_array
plt.imshow(image, cmap=plt.cm.gray)
plt.show()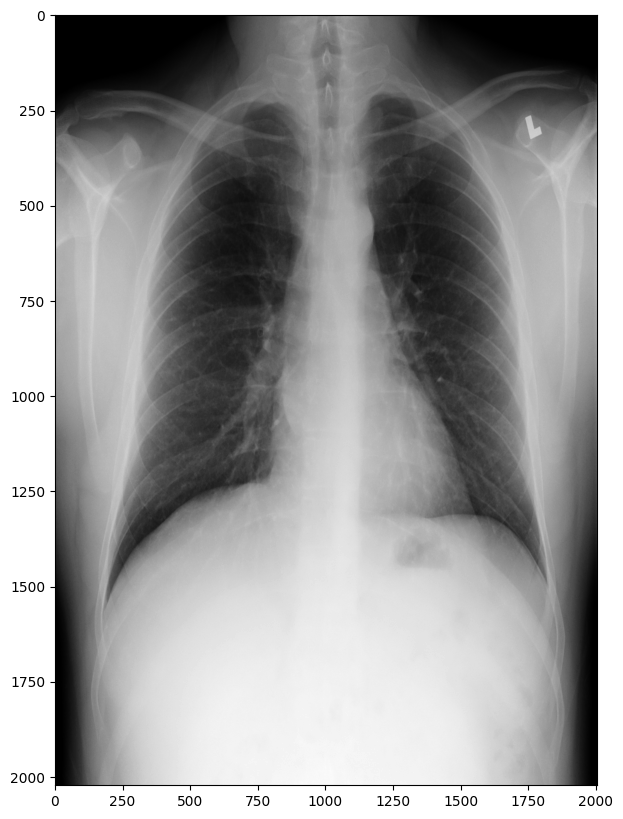
In this example, we use the pixel_array attribute to access the image data and store it in the image variable. We then use the imshow() function from the Matplotlib package to display the image.
Modifying DICOM Files
PyDICOM also provides a simple API for modifying DICOM files. You can modify metadata, image data, or both. Here is an example of how to modify the patient name in a DICOM file:
ds.PatientName = 'John Doe'
ds.save_as('new_image.dcm')In this example, we modify the patient name in the ds Dataset object and then save it as a new DICOM file named new_image.dcm. Note that you need to call the save_as() method to save the modified Dataset object as a new DICOM file.
In this article, we have explored the basics of working with DICOM files using PyDICOM. We have shown how to read DICOM files, access metadata and image data, and modify DICOM files. PyDICOM provides a powerful and flexible API for working with DICOM files, making it a great choice for medical image processing. For more details visits pydicom official documentation.
Comments (0)